News
May 2025: The grant to ImmGen is terminated by NIH.
Running on fumes.
March 2025: ImmGen welcomes ImmgenMaps!
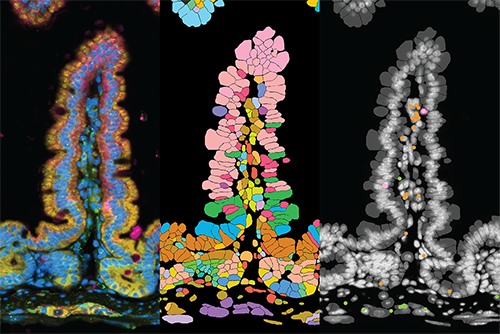 The ImmGenMaps project, led by the Reina lab, aims to generate a comprehensive atlas of immune cells across mouse organs in space.
Combining spatial gene expression profiling with detailed protein marker analysis, ImmGenMaps goes beyond traditional methods, capturing immune cells in their natural tissue context and preserving key spatial information.
The ImmGenMaps project, led by the Reina lab, aims to generate a comprehensive atlas of immune cells across mouse organs in space.
Combining spatial gene expression profiling with detailed protein marker analysis, ImmGenMaps goes beyond traditional methods, capturing immune cells in their natural tissue context and preserving key spatial information.
In its first phase, ImmGenMaps is generating high-quality datasets from 26 healthy mouse organs, establishing baseline maps for male and female specimens.
Future phases will explore immune responses under various challenges, including infections, cancer, allergies, and autoimmune conditions. ImmGenMaps invites researchers worldwide to contribute their expertise and interests via the usual ImmGen OpenSource collaborative format, ensuring that the resource is robust, versatile.
December 2024: ImmGen Network of the Year Announced

The ImmGen team has tallied the 100 most popular genes according to requests on the Skyline data browser (942 for Itgam) and displayed on a plot from the String database.
Dominant “Cyto/chemokine receptors” and “T cell differentiation” ontologies are showed in blue and red, respectively.
Best wishes for a Happy New Year to all ImmGen users!
December 2024

As usual, hardy ImmGen participants braved the delightful December weather in Boston (sub-freezing temps, this year) for the annual ImmGen Workshop. For the also usual mix of exciting results from ongoing programs, discussion of future directions, arguments over which is the most important immunological cell lineage.
And, of course, this year’s ImmGen T-shirt.
July 2024: GEM is retiring
 It is with mixed feelings that we announce the retirement of our Genome Expression Map (GEM) databrowser, which was the oldest of our viewers . Over the years, GEM provided a unique perspective into ImmGen data, allowing users to zoom in and out of the mouse genome.
It is with mixed feelings that we announce the retirement of our Genome Expression Map (GEM) databrowser, which was the oldest of our viewers . Over the years, GEM provided a unique perspective into ImmGen data, allowing users to zoom in and out of the mouse genome.
Because of changes in the GoogleMaps APIs, GEM had gone dark, and making/keeping it current had proven too demanding.
We have not abandoned the idea of using similar dynamics in the future, so that map aficionados may have their day again.
June 2024: Come explore the first data release from the immgenT program
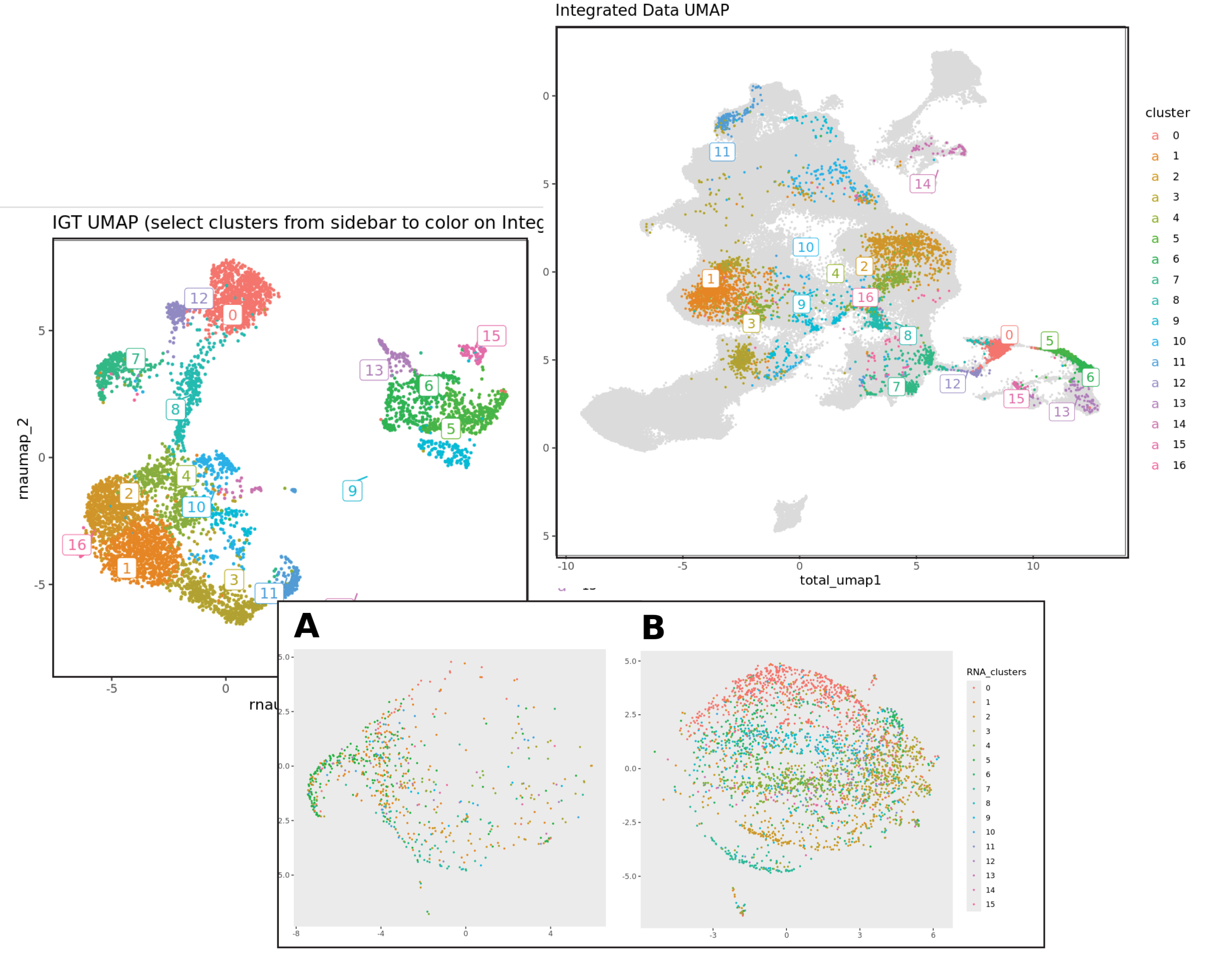 This Open Source Project is brought to you by the coordinated work of over 80 participants in 30 labs, aiming to profile every T cell in every body location of the mouse, and in every state that immunologic challenges push T cells into. Profiling was performed at the single-cell level, with the combined determination of mRNA, surface protein (CITE-seq/TotalSeq), and TCR-VDJ sequence (both chains). The CITE-Seq panel was designed in collaboration with BioLegend.
This Open Source Project is brought to you by the coordinated work of over 80 participants in 30 labs, aiming to profile every T cell in every body location of the mouse, and in every state that immunologic challenges push T cells into. Profiling was performed at the single-cell level, with the combined determination of mRNA, surface protein (CITE-seq/TotalSeq), and TCR-VDJ sequence (both chains). The CITE-Seq panel was designed in collaboration with BioLegend.
In line with ImmGen operating principles, the data are made publicly accessible pre-publication via a dedicated browser, Rosetta, developed by Dania Mallah, Niket Patel, and Brinda Vijaykumar. You can explore specific datasets, augmented by their integration in reference to other T cell datasets and a flow-like experience of surface marker expression. For more information about immgenT, and to stay updated on our latest advancements, visit immgenT
Go T!
May 2024: New Chromatin databrowser that maps the Histone code across the immune system
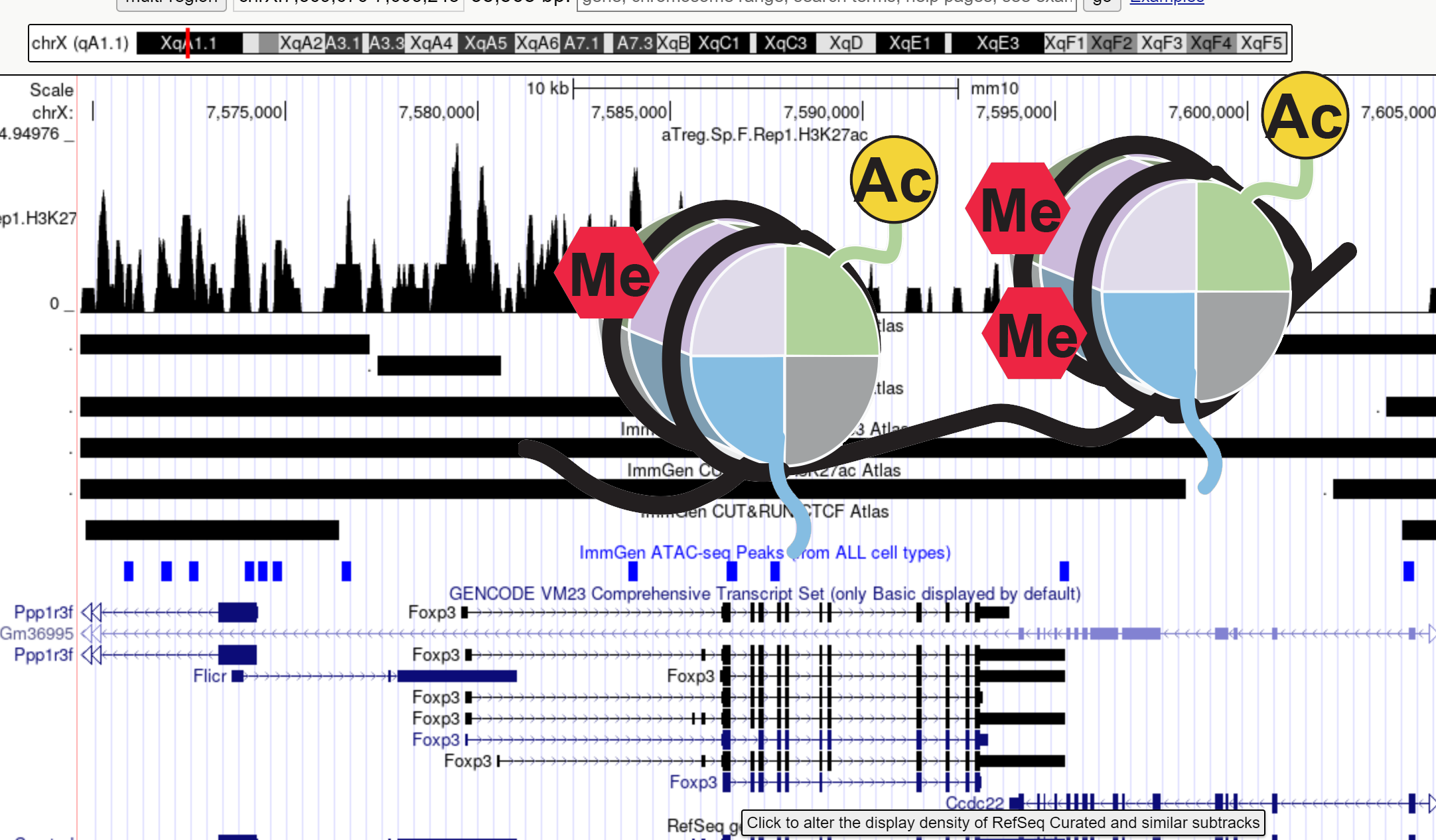 Immgen groups have performed a large-scale mapping of the main histone post-transcriptional modifications (“histone code”) across the immune system of the mouse, from mast cells to CD8+ memory T cells. CUT&RUN data, all from in primary cells ex-vivo. This large effort was managed by Juliana Lee (ImmGen coordinator) and Dughan Ahimovic (Josefowicz lab), and was a tight collaboration with Epicypher, the leading Epigenomics company, who implemented a low-input automated variant of the CUT&RUN technology necessary to process the low cell numbers available from our narrow sort selections. Fourteen investigators from 11 ImmGen labs were involved in the sample preparation, yielding 1126 QC-passing samples from 94 different cell types (both males and females represented).
Immgen groups have performed a large-scale mapping of the main histone post-transcriptional modifications (“histone code”) across the immune system of the mouse, from mast cells to CD8+ memory T cells. CUT&RUN data, all from in primary cells ex-vivo. This large effort was managed by Juliana Lee (ImmGen coordinator) and Dughan Ahimovic (Josefowicz lab), and was a tight collaboration with Epicypher, the leading Epigenomics company, who implemented a low-input automated variant of the CUT&RUN technology necessary to process the low cell numbers available from our narrow sort selections. Fourteen investigators from 11 ImmGen labs were involved in the sample preparation, yielding 1126 QC-passing samples from 94 different cell types (both males and females represented).
These data are being analyzed by several participants and publications should be forthcoming, but per ImmGen practice the data are being made available for public browsing on a dedicated databrowser implemented by Liang Yang. Select one or more samples by lineage, cell type and modification (H3K4me1, H3K4me3, H3K27Ac, H3K27me3, H3K36me3, and CTCF), then choose a gene or a genomic interval to visualize feature peaks within the region.
Sample preparation and analysis: Michela Frascoli, Enxhi Ferraj, Sara Quon, Kennidy Takehara, Tamara Salloum, Alberto Milian, Natalia Jaeger, Vincent Peng, Mallory Paynich, Samarth Hegde, Julie Tellier, Marc Todd, Jonathan Kotzin, Gabriel Ascui-Gac, Fabiana Duarte, and Juliana Lee in the Kang, Goldrath, Dwyer, Lanier, Colonna, Kronenberg, Merad, Nutt, Nigrovic and CBDM labs.
Computational Analysis team (alphabetical): Dughan Ahimovic, Michael Bale, Andrew Earl, Kaili Fan, Laszlo Halasz, Alexander Sasse, Brian Soong, Anna Spiro, Christophe Benoist, Jason Buenrostro, Steve Josefowicz, Sara Mostafavi, Steve Nutt.
February 2024: Check out the new ImmGen forum!

It’s now easy to share ideas, look up answers to FAQs, or ask questions to the ImmGen team or the community of ImmGen data users.
Thanks for registering your email to add posts and comment on previous ones (solely to avoid spam and junk).
November 2023: ImmGen is powered by ChatGPT.

(In a small part)
Artificial Intelligence now informs ImmGen’s databrowsers. Not the expression profiles or epigenomic data, which are still experimentally-determined. But, in an effort to increase the information served about genes or gene clusters and what they might mean, OpenAI’s ChatGPT 3.5 will be used to query the function of a gene, or relationships between genes in the databrowsers, with answers that can be more immediately intuitive and integrated than those delivered by the usual tools.
Disclaimer: ChatGPT’s answers are not always completely true (and it may plead ignorance in some cases).
So “caveat emptor”, but these functions represent the start of ImmGen’s interest in including AI and Machine Learning to enhance the knowledge it serves.
August 2023: New transcriptional signatures to inflammatory Cytokines.
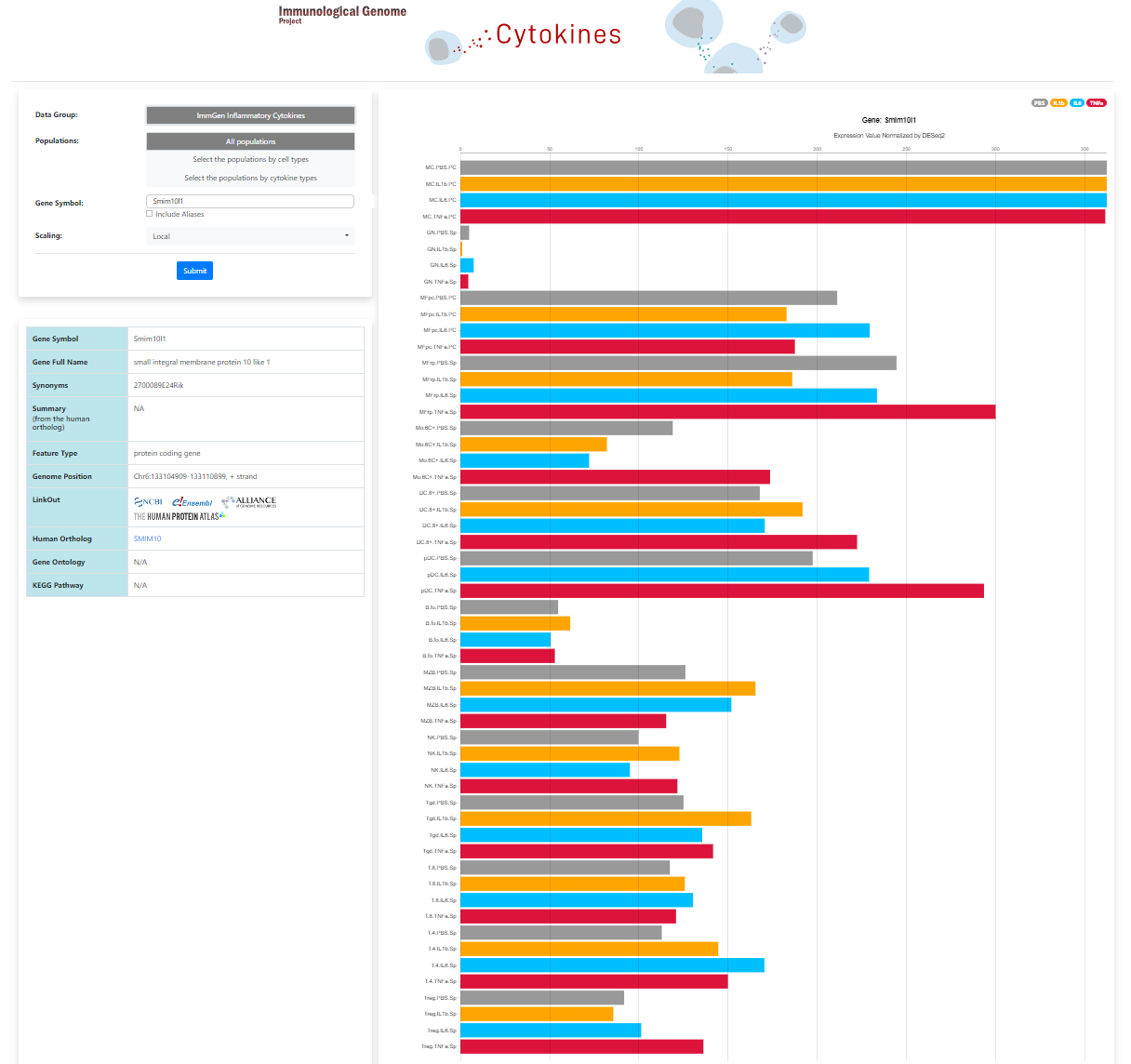
In the latest installment of ImmGen’s goal of systematically determining the transcriptional signatures of all cytokines across all immunologic lineages, Juliana Lee has generated gene expression profiles (RNAseq on carefully sorted cell populations) of the responses to key inflammatory cytokines (IL1-beta, Il6, and TNF-alpha) across the ImmGen standard 14-cell-set.
Use the Skyline databrowser to display cytokine-induced expression profiles across the ImmGen immunological cell-types for a selected gene, or download signature sets for each cytokine.
June 2023: Human data in the ImmGen app
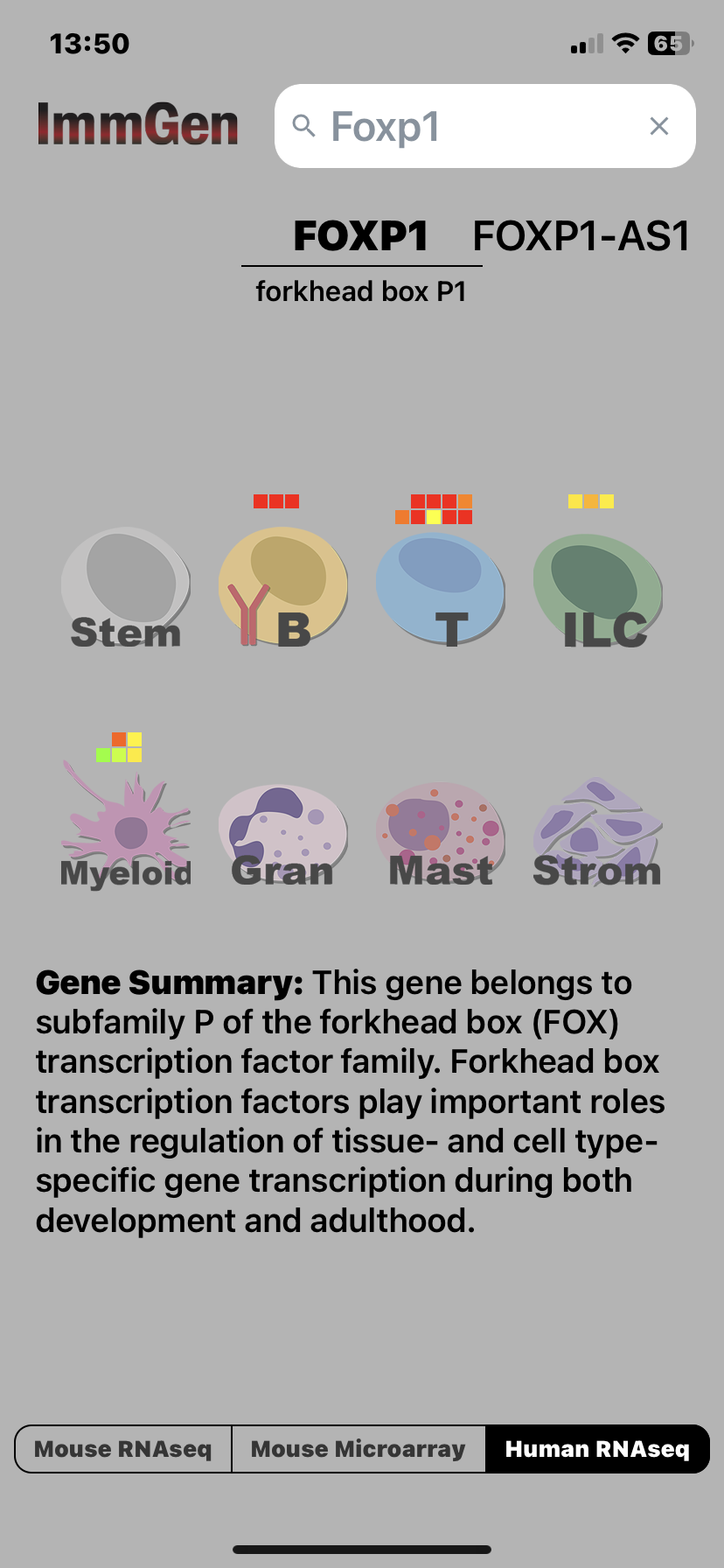
ImmGen developer Jeff Ericson has updated the ImmGen Mobile app, adding a Human RNAseq datagroup. The new release of the app displays human RNAseq data from the main populations of human PBMCs, in addition to the previous mouse data. Search for a Gene Symbol, and the app displays gene expression levels in different lineages of the immune system, toggle between human and mouse. Updated Gene Summaries from NCBI have been added. Download the new Version from the Apple iTunes Store or the Google Play store using the keyword “ImmGen”. This update is automatically applied on existing installs.
The ImmGen “Populations” list is ever growing !
Immune cell types have been isolated and added to ImmGen’s already long population collection of flow-purified.
The new additions include B cells (Julie Tellier, Nutt Lab), activated and memory CD8 T (Ty Crowl, Goldrath lab), MAIT (Mallory Paynich, Kronenberg lab) and more Mast cells than most Immunologists ever knew existed (Tamara Salloum, Dwyer lab).
You can now browse and compare their expression profiles with ImmGen’s most popular databrowsers : Gene Skyline, Population Comparison and MyGeneSet.
“AlwaysOpen” ImmGen
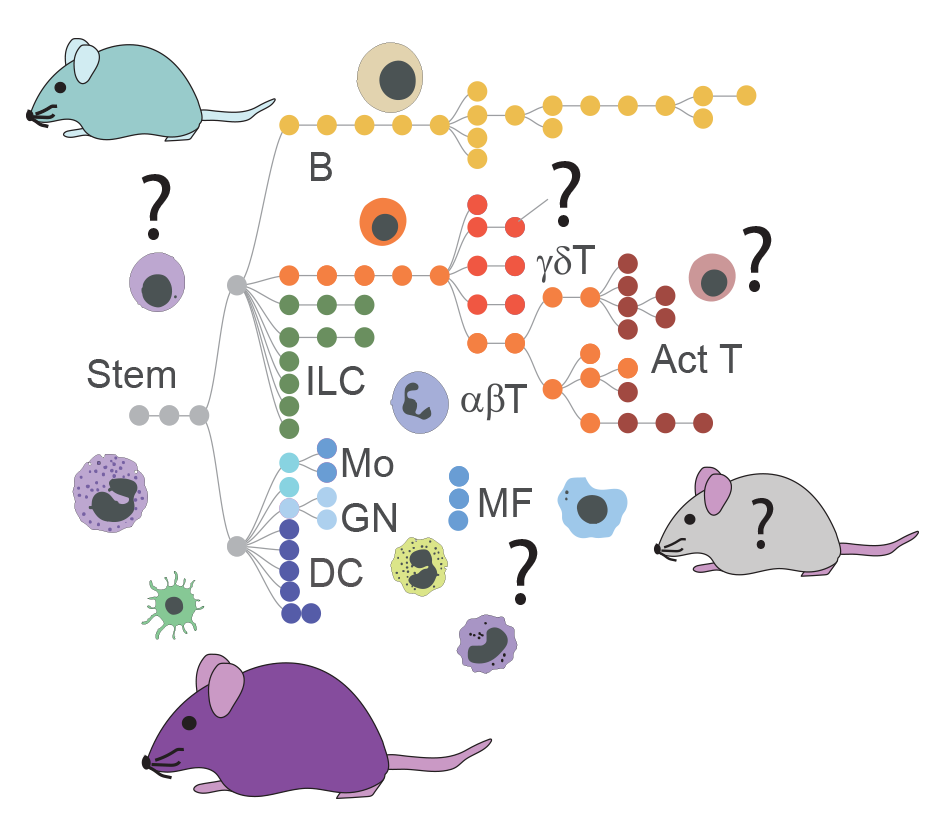
ImmGen is now launching a new “AlwaysOpen” initiative, to open its workhorse low-input RNAseq pipeline to any qualified laboratory interested in helping to generate RNAseq datasets that fit their own interests, while contributing to the compendium displayed on the ImmGen website and mobile apps.
Yes, free profiling.
That being said, ImmGen is not turning into a core facility. Proposals needs to meet criteria of general interest, and sample preparation will need to follow obsessively the ImmGen SOP.
More details
Dec 2022: Our ImmGen app gets a new feature!
ImmGen developer Jeff Ericson has updated the ImmGen Mobile up, bringing the datasets to speed and adding a mobile version of the Gene Constellation databrowser.
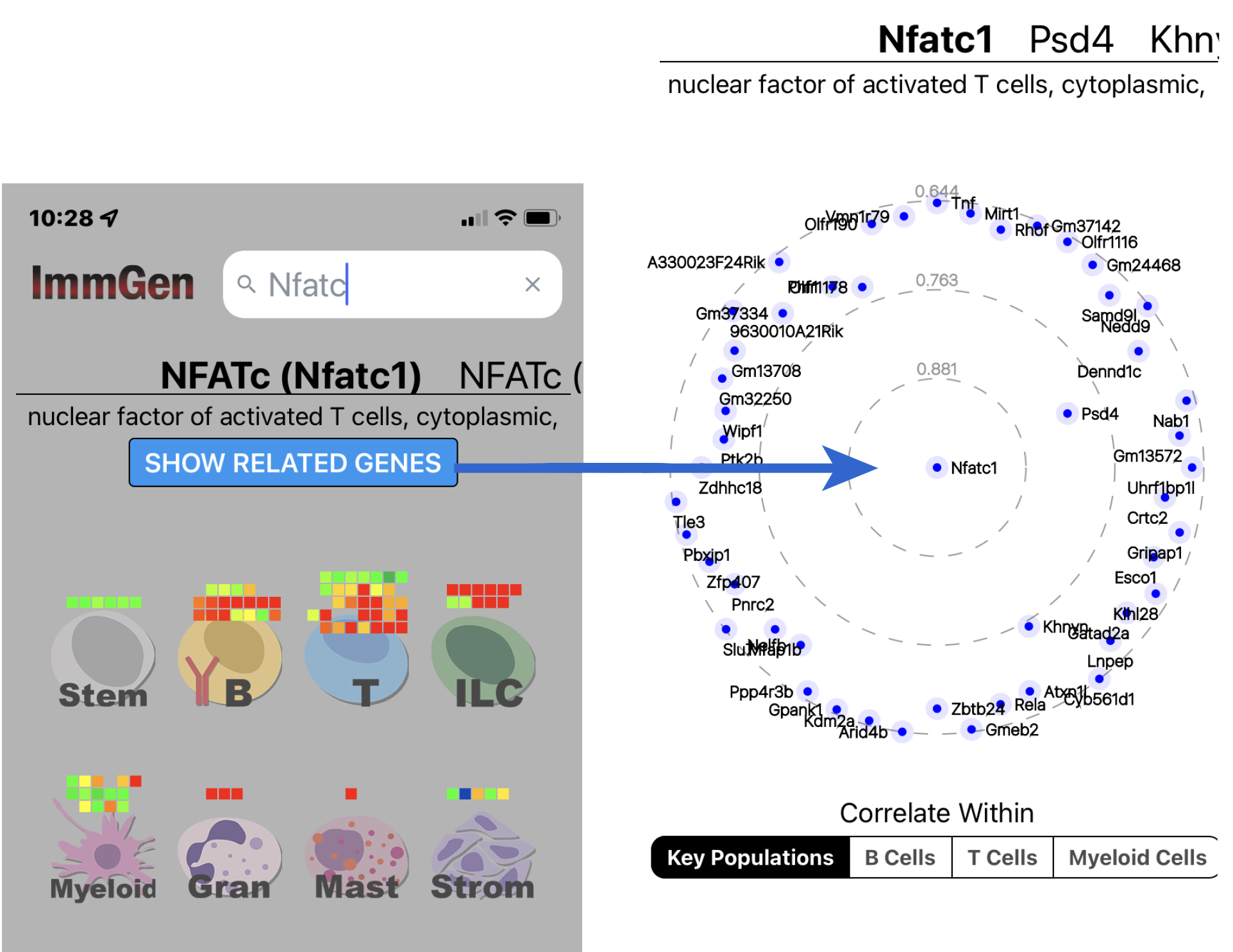 As in previous ImmGen app versions, search for a Gene Symbol, and the app displays gene expression levels in different lineages of the mouse immune system, either as 2-D barcode view for all lineages or as detailed histograms for specific lineages.
The new constellation feature allows users to visualize correlation networks for a chosen gene, showing the genes whose expression within a selected lineage of immunocytes is most related to the queried gene (and, on rainy afternoons, can iteratively look for genes most related one of those genes, and so on)
As in previous ImmGen app versions, search for a Gene Symbol, and the app displays gene expression levels in different lineages of the mouse immune system, either as 2-D barcode view for all lineages or as detailed histograms for specific lineages.
The new constellation feature allows users to visualize correlation networks for a chosen gene, showing the genes whose expression within a selected lineage of immunocytes is most related to the queried gene (and, on rainy afternoons, can iteratively look for genes most related one of those genes, and so on)
Deployed simultaneously on both Android and IOS apps, the new release of the app displays RNAseq data as well as previous microarray data, easily switched through a toggle button. Download Version 3.1.0 from the Apple iTunes Store or the Google Play store using the keyword “ImmGen”. Future updates will be applied automatically on existing apps.
May 2022: ImmGen launches a new Open Source Project!
Per the recent Correspondence in NatImm, ImmGen launches a new Open Source Project! After the success of the OpenSource MNP, when myeloid cells had their day, time for the real deal: T cells! They have been named killers, helpers. But who are they really?
The goal is to coordinate wide community participation (beyond the core Immgen labs) around a simple project goal: to profile every T cell, in every body location of the mouse, and in every state that immunologic challenges push T cells into. You name it and we’ll sequence it (TM)!
• T cells across body locations (e.g. lymphoid and parenchymal organs): check
• bacterial, fungal, viral and parasitic infections, autoimmune and inflammatory lesions, tumors, pharmacological intervention: check
• Over time, sex and genetic background: check
Profiling will be performed at the single-cell level, with the combined measurement of mRNA, surface protein (CITE-seq/TotalSeq) expression, and TCR-V sequencing (both chains, real clonotypes). The ImmGen core team collaborated with BioLegend to define, test and optimize a panel of DNA-coded antibodies to proteins on the surfaces of T cells. These will allow a direct connection between RNA and protein content, resolving cell-types/states from both the RNA and protein side (jointly displayed on the Rosetta viewer).
This OpenSource Project is open to all labs. If you have an idea, a specific know-how, a model of T cell function/challenge that you feel is in scope (see Forum), and whose workup you would like to be actively involved in, please consider participating by submitting a short proposal and justification.
December 2021: Immgen Gene of the Year Announced

The ImmGen team has tallied the number of times each GeneSymbol was requested during 2021 on the skyline databrowser.
The winner is Il7r, requested 300 times over the course of the year, followed by 3 very close contenders: Itgax (228 times), Il2ra (227 times) and Tcf7 (226 times).
Best wishes for a Happy New Year to all ImmGen users!
Running on fumes.
March 2025: ImmGen welcomes ImmgenMaps!

 The ImmGenMaps project, led by the Reina lab, aims to generate a comprehensive atlas of immune cells across mouse organs in space.
Combining spatial gene expression profiling with detailed protein marker analysis, ImmGenMaps goes beyond traditional methods, capturing immune cells in their natural tissue context and preserving key spatial information.
The ImmGenMaps project, led by the Reina lab, aims to generate a comprehensive atlas of immune cells across mouse organs in space.
Combining spatial gene expression profiling with detailed protein marker analysis, ImmGenMaps goes beyond traditional methods, capturing immune cells in their natural tissue context and preserving key spatial information.In its first phase, ImmGenMaps is generating high-quality datasets from 26 healthy mouse organs, establishing baseline maps for male and female specimens.
Future phases will explore immune responses under various challenges, including infections, cancer, allergies, and autoimmune conditions. ImmGenMaps invites researchers worldwide to contribute their expertise and interests via the usual ImmGen OpenSource collaborative format, ensuring that the resource is robust, versatile.
December 2024: ImmGen Network of the Year Announced

The ImmGen team has tallied the 100 most popular genes according to requests on the Skyline data browser (942 for Itgam) and displayed on a plot from the String database.
Dominant “Cyto/chemokine receptors” and “T cell differentiation” ontologies are showed in blue and red, respectively.
Best wishes for a Happy New Year to all ImmGen users!
December 2024

As usual, hardy ImmGen participants braved the delightful December weather in Boston (sub-freezing temps, this year) for the annual ImmGen Workshop. For the also usual mix of exciting results from ongoing programs, discussion of future directions, arguments over which is the most important immunological cell lineage.
And, of course, this year’s ImmGen T-shirt.
July 2024: GEM is retiring
 It is with mixed feelings that we announce the retirement of our Genome Expression Map (GEM) databrowser, which was the oldest of our viewers . Over the years, GEM provided a unique perspective into ImmGen data, allowing users to zoom in and out of the mouse genome.
It is with mixed feelings that we announce the retirement of our Genome Expression Map (GEM) databrowser, which was the oldest of our viewers . Over the years, GEM provided a unique perspective into ImmGen data, allowing users to zoom in and out of the mouse genome.Because of changes in the GoogleMaps APIs, GEM had gone dark, and making/keeping it current had proven too demanding.
We have not abandoned the idea of using similar dynamics in the future, so that map aficionados may have their day again.
June 2024: Come explore the first data release from the immgenT program
 This Open Source Project is brought to you by the coordinated work of over 80 participants in 30 labs, aiming to profile every T cell in every body location of the mouse, and in every state that immunologic challenges push T cells into. Profiling was performed at the single-cell level, with the combined determination of mRNA, surface protein (CITE-seq/TotalSeq), and TCR-VDJ sequence (both chains). The CITE-Seq panel was designed in collaboration with BioLegend.
This Open Source Project is brought to you by the coordinated work of over 80 participants in 30 labs, aiming to profile every T cell in every body location of the mouse, and in every state that immunologic challenges push T cells into. Profiling was performed at the single-cell level, with the combined determination of mRNA, surface protein (CITE-seq/TotalSeq), and TCR-VDJ sequence (both chains). The CITE-Seq panel was designed in collaboration with BioLegend.In line with ImmGen operating principles, the data are made publicly accessible pre-publication via a dedicated browser, Rosetta, developed by Dania Mallah, Niket Patel, and Brinda Vijaykumar. You can explore specific datasets, augmented by their integration in reference to other T cell datasets and a flow-like experience of surface marker expression. For more information about immgenT, and to stay updated on our latest advancements, visit immgenT
Go T!
May 2024: New Chromatin databrowser that maps the Histone code across the immune system
 Immgen groups have performed a large-scale mapping of the main histone post-transcriptional modifications (“histone code”) across the immune system of the mouse, from mast cells to CD8+ memory T cells. CUT&RUN data, all from in primary cells ex-vivo. This large effort was managed by Juliana Lee (ImmGen coordinator) and Dughan Ahimovic (Josefowicz lab), and was a tight collaboration with Epicypher, the leading Epigenomics company, who implemented a low-input automated variant of the CUT&RUN technology necessary to process the low cell numbers available from our narrow sort selections. Fourteen investigators from 11 ImmGen labs were involved in the sample preparation, yielding 1126 QC-passing samples from 94 different cell types (both males and females represented).
Immgen groups have performed a large-scale mapping of the main histone post-transcriptional modifications (“histone code”) across the immune system of the mouse, from mast cells to CD8+ memory T cells. CUT&RUN data, all from in primary cells ex-vivo. This large effort was managed by Juliana Lee (ImmGen coordinator) and Dughan Ahimovic (Josefowicz lab), and was a tight collaboration with Epicypher, the leading Epigenomics company, who implemented a low-input automated variant of the CUT&RUN technology necessary to process the low cell numbers available from our narrow sort selections. Fourteen investigators from 11 ImmGen labs were involved in the sample preparation, yielding 1126 QC-passing samples from 94 different cell types (both males and females represented).These data are being analyzed by several participants and publications should be forthcoming, but per ImmGen practice the data are being made available for public browsing on a dedicated databrowser implemented by Liang Yang. Select one or more samples by lineage, cell type and modification (H3K4me1, H3K4me3, H3K27Ac, H3K27me3, H3K36me3, and CTCF), then choose a gene or a genomic interval to visualize feature peaks within the region.
Sample preparation and analysis: Michela Frascoli, Enxhi Ferraj, Sara Quon, Kennidy Takehara, Tamara Salloum, Alberto Milian, Natalia Jaeger, Vincent Peng, Mallory Paynich, Samarth Hegde, Julie Tellier, Marc Todd, Jonathan Kotzin, Gabriel Ascui-Gac, Fabiana Duarte, and Juliana Lee in the Kang, Goldrath, Dwyer, Lanier, Colonna, Kronenberg, Merad, Nutt, Nigrovic and CBDM labs.
Computational Analysis team (alphabetical): Dughan Ahimovic, Michael Bale, Andrew Earl, Kaili Fan, Laszlo Halasz, Alexander Sasse, Brian Soong, Anna Spiro, Christophe Benoist, Jason Buenrostro, Steve Josefowicz, Sara Mostafavi, Steve Nutt.
February 2024: Check out the new ImmGen forum!
It’s now easy to share ideas, look up answers to FAQs, or ask questions to the ImmGen team or the community of ImmGen data users.
Thanks for registering your email to add posts and comment on previous ones (solely to avoid spam and junk).
November 2023: ImmGen is powered by ChatGPT.

(In a small part)
Artificial Intelligence now informs ImmGen’s databrowsers. Not the expression profiles or epigenomic data, which are still experimentally-determined. But, in an effort to increase the information served about genes or gene clusters and what they might mean, OpenAI’s ChatGPT 3.5 will be used to query the function of a gene, or relationships between genes in the databrowsers, with answers that can be more immediately intuitive and integrated than those delivered by the usual tools.
Disclaimer: ChatGPT’s answers are not always completely true (and it may plead ignorance in some cases).
So “caveat emptor”, but these functions represent the start of ImmGen’s interest in including AI and Machine Learning to enhance the knowledge it serves.
August 2023: New transcriptional signatures to inflammatory Cytokines.

In the latest installment of ImmGen’s goal of systematically determining the transcriptional signatures of all cytokines across all immunologic lineages, Juliana Lee has generated gene expression profiles (RNAseq on carefully sorted cell populations) of the responses to key inflammatory cytokines (IL1-beta, Il6, and TNF-alpha) across the ImmGen standard 14-cell-set.
Use the Skyline databrowser to display cytokine-induced expression profiles across the ImmGen immunological cell-types for a selected gene, or download signature sets for each cytokine.
June 2023: Human data in the ImmGen app

ImmGen developer Jeff Ericson has updated the ImmGen Mobile app, adding a Human RNAseq datagroup. The new release of the app displays human RNAseq data from the main populations of human PBMCs, in addition to the previous mouse data. Search for a Gene Symbol, and the app displays gene expression levels in different lineages of the immune system, toggle between human and mouse. Updated Gene Summaries from NCBI have been added. Download the new Version from the Apple iTunes Store or the Google Play store using the keyword “ImmGen”. This update is automatically applied on existing installs.
The ImmGen “Populations” list is ever growing !

Immune cell types have been isolated and added to ImmGen’s already long population collection of flow-purified.
The new additions include B cells (Julie Tellier, Nutt Lab), activated and memory CD8 T (Ty Crowl, Goldrath lab), MAIT (Mallory Paynich, Kronenberg lab) and more Mast cells than most Immunologists ever knew existed (Tamara Salloum, Dwyer lab).
You can now browse and compare their expression profiles with ImmGen’s most popular databrowsers : Gene Skyline, Population Comparison and MyGeneSet.
“AlwaysOpen” ImmGen

ImmGen is now launching a new “AlwaysOpen” initiative, to open its workhorse low-input RNAseq pipeline to any qualified laboratory interested in helping to generate RNAseq datasets that fit their own interests, while contributing to the compendium displayed on the ImmGen website and mobile apps.
Yes, free profiling.
That being said, ImmGen is not turning into a core facility. Proposals needs to meet criteria of general interest, and sample preparation will need to follow obsessively the ImmGen SOP.
More details
Dec 2022: Our ImmGen app gets a new feature!
ImmGen developer Jeff Ericson has updated the ImmGen Mobile up, bringing the datasets to speed and adding a mobile version of the Gene Constellation databrowser.
 As in previous ImmGen app versions, search for a Gene Symbol, and the app displays gene expression levels in different lineages of the mouse immune system, either as 2-D barcode view for all lineages or as detailed histograms for specific lineages.
The new constellation feature allows users to visualize correlation networks for a chosen gene, showing the genes whose expression within a selected lineage of immunocytes is most related to the queried gene (and, on rainy afternoons, can iteratively look for genes most related one of those genes, and so on)
As in previous ImmGen app versions, search for a Gene Symbol, and the app displays gene expression levels in different lineages of the mouse immune system, either as 2-D barcode view for all lineages or as detailed histograms for specific lineages.
The new constellation feature allows users to visualize correlation networks for a chosen gene, showing the genes whose expression within a selected lineage of immunocytes is most related to the queried gene (and, on rainy afternoons, can iteratively look for genes most related one of those genes, and so on)
Deployed simultaneously on both Android and IOS apps, the new release of the app displays RNAseq data as well as previous microarray data, easily switched through a toggle button. Download Version 3.1.0 from the Apple iTunes Store or the Google Play store using the keyword “ImmGen”. Future updates will be applied automatically on existing apps.
May 2022: ImmGen launches a new Open Source Project!

Per the recent Correspondence in NatImm, ImmGen launches a new Open Source Project! After the success of the OpenSource MNP, when myeloid cells had their day, time for the real deal: T cells! They have been named killers, helpers. But who are they really?
The goal is to coordinate wide community participation (beyond the core Immgen labs) around a simple project goal: to profile every T cell, in every body location of the mouse, and in every state that immunologic challenges push T cells into. You name it and we’ll sequence it (TM)!
• T cells across body locations (e.g. lymphoid and parenchymal organs): check
• bacterial, fungal, viral and parasitic infections, autoimmune and inflammatory lesions, tumors, pharmacological intervention: check
• Over time, sex and genetic background: check
Profiling will be performed at the single-cell level, with the combined measurement of mRNA, surface protein (CITE-seq/TotalSeq) expression, and TCR-V sequencing (both chains, real clonotypes). The ImmGen core team collaborated with BioLegend to define, test and optimize a panel of DNA-coded antibodies to proteins on the surfaces of T cells. These will allow a direct connection between RNA and protein content, resolving cell-types/states from both the RNA and protein side (jointly displayed on the Rosetta viewer).
This OpenSource Project is open to all labs. If you have an idea, a specific know-how, a model of T cell function/challenge that you feel is in scope (see Forum), and whose workup you would like to be actively involved in, please consider participating by submitting a short proposal and justification.
December 2021: Immgen Gene of the Year Announced

The ImmGen team has tallied the number of times each GeneSymbol was requested during 2021 on the skyline databrowser.
The winner is Il7r, requested 300 times over the course of the year, followed by 3 very close contenders: Itgax (228 times), Il2ra (227 times) and Tcf7 (226 times).
Best wishes for a Happy New Year to all ImmGen users!